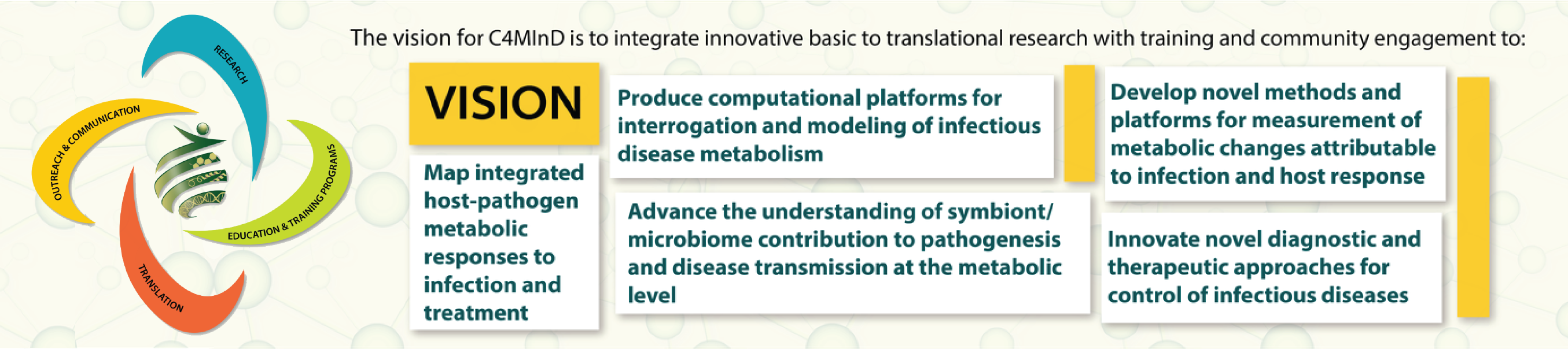
The Center for Metabolism of Infectious Diseases
Along with Dr. John Belisle, Dr. Rushika Perera is the Co-Director of the newly established Center for Metabolism of Infectious Diseases (C4MInD).C4MInD brings together the expertise of over 65 CSU researchers to enable development of new treatments, preventions and diagnostics for infectious diseases by resolving host-vector-pathogen-environment interactions at a metabolic level.
COVID-19: Developing platforms to test antivirals against SARS-CoV-2
The Perera lab (in collaboration with the Geiss Laboratory and the CSU Office of the Vice President for Research) has mobilized testing of FDA-approved antiviral drugs that can be repurposed and rapidly driven into clinical trials, as well as new compounds that can be evaluated for efficacy against SARS-CoV-2. We are currently working with collaborators and partners from around the world to screen their drug and compound inventories.
In collaboration with colleagues in the Center for Metabolism of Infectious Diseases (C4MInD), we are also investigating how metabolic pathways altered in chronic diseases such as diabetes, metabolic syndrome, heart disease and obesity might alter the efficacy of therapeutics against SARS-CoV-2. In this work, we are in investigating how host targeted therapies can be combined with known antagonists of viral replicase enzymes to provide synergistic binary and tertiary combinations of therapeutics that have greater efficacy than the monotherapies alone. This work includes evaluating synergistic, additive and antagonistic interactions between FDA approved drugs as well as investigational compounds.
Metabolite biomarkers of severe disease, resolution and therapeutic efficacy
It is currently impossible to differentiate between patients with dengue disease, who will have an unremarkable disease episode from those who will progress to severe disease and perhaps death. There are no standardized prognostic biomarkers for disease outcome. We have developed metabolite biosignatures that can differentiate dengue fever (DF), dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS) from each other and from infections with Chikungunya and Zika virus or non-dengue febrile diseases. Metabolite biosignatures can be utilized for early detection and triaging of dengue patients, to assist in better clinical management of those at greater risk. They can also be utilized to identify disease resolution, recurrence and/or therapeutic efficacy. The long-term goal of this effort is to develop similar biosignatures for saliva and urine from patients such that non-invasive point-of-care (POC) tests are developed for diagnosis and prognosis (D&P) of dengue disease and therapeutic efficacy.
Collaborators
Barry Beaty, Ph.D.
Carol Blair, Ph.D.
Center for Vector-Borne Infectious Diseases, Colorado State University
Eva Harris, Ph.D.
Division of Infectious Diseases and Vaccinology, School of Public Health, University of California, Berkeley, CA
Sustainable Sciences Institute, Managua, Nicaragua
Angel Balmaseda, M.D.
Laboratorio Nacional de Virología, Centro Nacional de Diagnóstico y Referencia, Ministry of Health, Managua, Nicaragua
Metabolic basis of mosquito-endosymbiont-virus interactions
These studies will identify biochemical pathways that change in the mosquito following infection with arboviruses, and how the endosymbiont Wolbachia used to control virus transmission in Aedes aegypti may be metabolically competing with the virus. Identifying these pathways that are integral to mosquito biology, provide a novel avenue to interfere with vector transmission of the virus.
Collaborators
Elizabeth McGraw, Ph.D.
Jason Rasgon, Ph.D.
Pennsylvania State University
Exploiting vulnerabilities in mosquito metabolism for prevention of human arboviral transmissions
Zika, dengue, chikungunya and yellow fever viruses are spread by the same mosquito vector, Aedes aegypti. The Perera lab studies specific metabolic processes in mosquitos required for successful viral replication. These studies will identify metabolic “choke-points” that can be exploited to develop interventions and thus block mosquito-human viral transmission.
Mosquito gut may hold the key to preventing Dengue and Zika – Purdue University [03/06/2018]
Metabolic basis of insecticide resistance
We are investigating the metabolic basis of insecticide resistance in Aedes aegypti. We have developed capability to trace the metabolites of insecticides and measure the enzymatic activities responsible for insecticide break down and development of resistance. These studies will assist in driving improvements in insecticide design and informed use of insecticides that are more effective on specific geographically isolated populations of vectors.
Collaborators
William Black IV, Ph.D.
Karla Saavedra, Ph.D.
Center for Vector-Borne Infectious Diseases, Colorado State University
Americo Rodriguez, Ph.D.
Rosa Patricia Penilla Navarro, Ph.D.
Centro Regional de Investigación en Salud Pública (Regional Center for Research in Public Health), Chiapas, Mexico
Harnessing the power of natural products as chemical synergists for insecticides.
We are investigating the properties of crude extracts and natural products endemic to Jamaica as novel insecticides or chemical synergists for currently used insecticides. These studies are also analyzing the metabolic impact of these compounds on mosquito-virus interactions and virus transmission. The impact of climate change on mosquito metabolism, vector competence, and the efficacy of insecticides are also being evaluated.
Collaborators
Thejani Rupika Delgoda, Ph.D.
Sheena Francis, Ph.D.
Natural Products Institute
University of the West Indies, Mona, Jamaica
Flavivirus manipulation of host metabolism: Mechanisms and Pathogenic Implications
Lipid metabolism is a fundamental cellular process that is hijacked by viruses to benefit their replicative needs. We are employing a multidisciplinary approach including single cell analyses, mass spectrometry, lipid biochemistry, conventional molecular and cellular biology to interrogate the molecular mechanism and viral components by which positive strand RNA viruses modulate host metabolic pathways to alter the host lipid repertoire for a replicative advantage.